Recently, a new study by Prof. Kenichi Tsuda and his team from the State Key Laboratory of Agricultural Microbiology and the College of Plant Science and Technology reveals the evolutionary mechanism of the PTI immune response in cruciferous plants.
During the long-term evolution of plants, the variation of stress response genes is of importance for plants to adapt to environmental changes. However, it is not clear that the evolutionary mechanism of gene transcription in plants under stress. Plants have evolved membrane-localized pattern recognition receptors (PRRs) to recognize conserved microbial-associated molecular patterns (MAMPs) on the surface of pathogenic microorganisms, and to activate pattern-induced immune responses (PTI). For example, FLG22, an oligopeptide at the N-terminal of bacterial flagellin, can be recognized by the Arabidopsis receptor FLS2 and trigger a series of PTI immune responses,including MAPK phosphorylation, transcriptional reprogramming, and hormone synthesis, thereby enhancing plant resistance to pathogens.At present, the understanding of the evolutionary mechanism of PTI is only limited to the evolution of PRR, and the conserved immune response and evolutionary mechanism of PTI among different plants are still poorly understood.
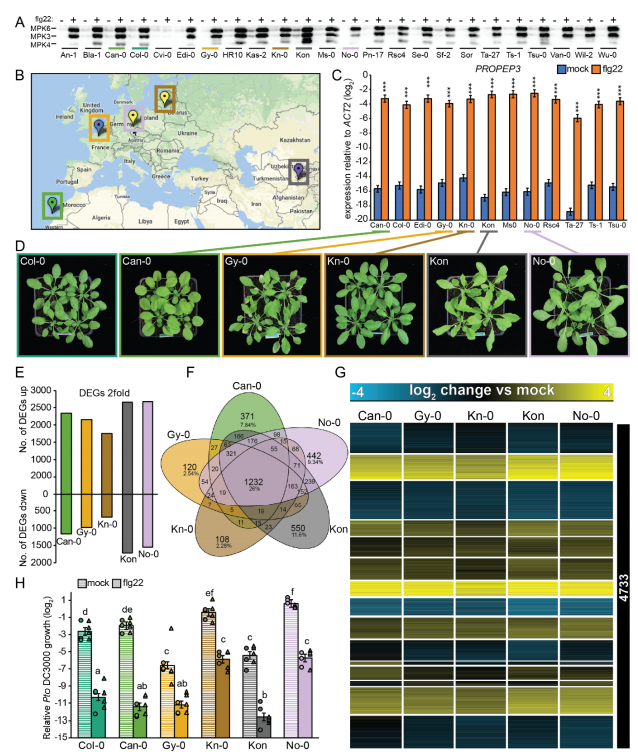
The researchers used flg22 treatment six arabidopsis lines and three kinds of cruciferae relatives (including shepherd 's-purse Capsella rubella, broken rice wil Cardamine hirsuta mustard and salt Eutrema salsugineum), found that four kinds of cruciferae plants can induction flg22 and trigger PTI early reaction, but flg22 PTI induced reaction caused by plant growth inhibition and resistance of pathogenic bacteria have obvious differences between the four kinds of plants.
To investigate the transcriptional evolution of the PTI immune response, researchers compared the transcriptome of six Arabidopsis strains and three cruciferous plants treated with FLG22. They found that there were some differentially expressed genes (DEG) and a considerable number of species-specific DEG in the four cruciferous plants.However, the responses of six Arabidopsis strains with different genetic backgrounds and geographical origins to FLG22 were highly conserved. However, differences in response to FLG22 among Cruciferae species are not consistent with their phylogeny.However, differences in response to FLG22 among Cruciferae species are not consistent with their phylogeny. They further found that WRKY transcription factor binding sites are enriched in the 5 '-cis-regulatory region of the species-specific FLG22 response gene, which may drive the evolution of species-specific gene transcription. Meanwhile, they also found that there was no correlation between coding region differences and transcriptome differences, and that purification and selection may maintain the conservatism of the common 5 '- regulatory region of the FLG22 response gene in cruciferous plants. In addition, the results showed that the metabolome of different Cruciferae treated with FLG22 was species-specific.
"The findings of this study shed light on the evolutionary mechanism of gene transcription during PTI in cruciferous plants and provide clues to the evolution of transcriptional levels in plants under biological stress." the researchers said.
Translated by: Wang Mengli
Source from: http://news.hzau.edu.cn/2021/0308/59506.shtml
Abstract:
Plants recognize surrounding microbes by sensing microbe-associated molecular patterns (MAMPs) to activate pattern-triggered immunity (PTI). Despite their significance for microbial control, the evolution of PTI responses remains largely uncharacterized. Here, by employing comparative transcriptomics of six Arabidopsis thaliana accessions and three additional Brassicaceae species to investigate PTI responses, we identified a set of genes that commonly respond to the MAMP flg22 and genes that exhibit species-specific expression signatures. Variation in flg22-triggered transcriptome responses across Brassicaceae species was incongruent with their phylogeny, while expression changes were strongly conserved within A. thaliana. We found the enrichment of WRKY transcription factor binding sites in the 5’-regulatory regions of conserved and species-specific responsive genes, linking the emergence of WRKY-binding sites with the evolution of gene expression patterns during PTI. Our findings advance our understanding of the evolution of the transcriptome during biotic stress.
Paper link:https://academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koab073/6159620?searchresult=1